Research
-
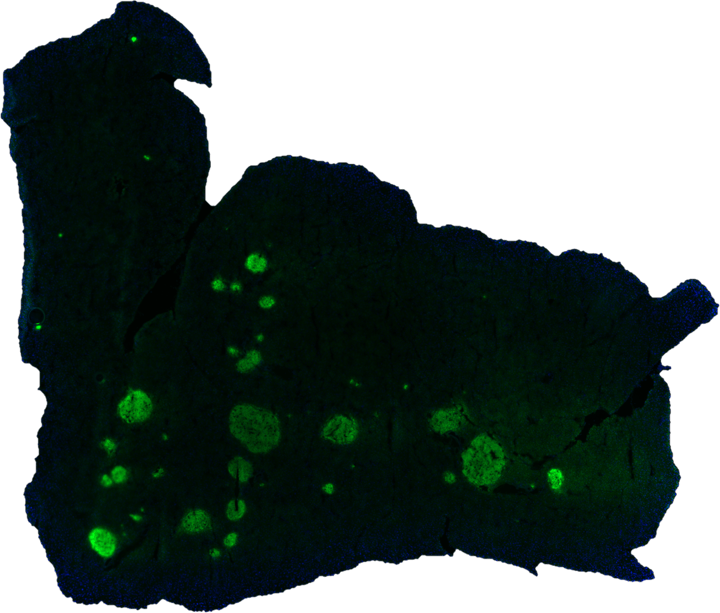 Mouse pancreas section stained with insulin.
Mouse pancreas section stained with insulin.Mass Spectrometry Imaging for Measuring in vivo Targeted Drug Delivery
Stanford University School of Medicine, 2018 - December 2020
Mentor(s): Dr. Justin Annes
Collaborator(s): Dr. Richard Zare, Sarah Noll
Mass spectrometry imaging (MSI) is a powerful technique to characterize spatial concentration of metabolites, small molecules, and proteins. We are developing desorption electrospray ionization (DESI)-MSI methods towards the following goals:
- Track in vivo drug distribution to characterize the efficacy of targeted drug delivery strategies.
- Characterize metabolic profile of pancreas and adrenal sections from different conditions (diabetes, age, etc.). These metabolic profiles will help us identify biochemical pathways involved in disease progression.
-
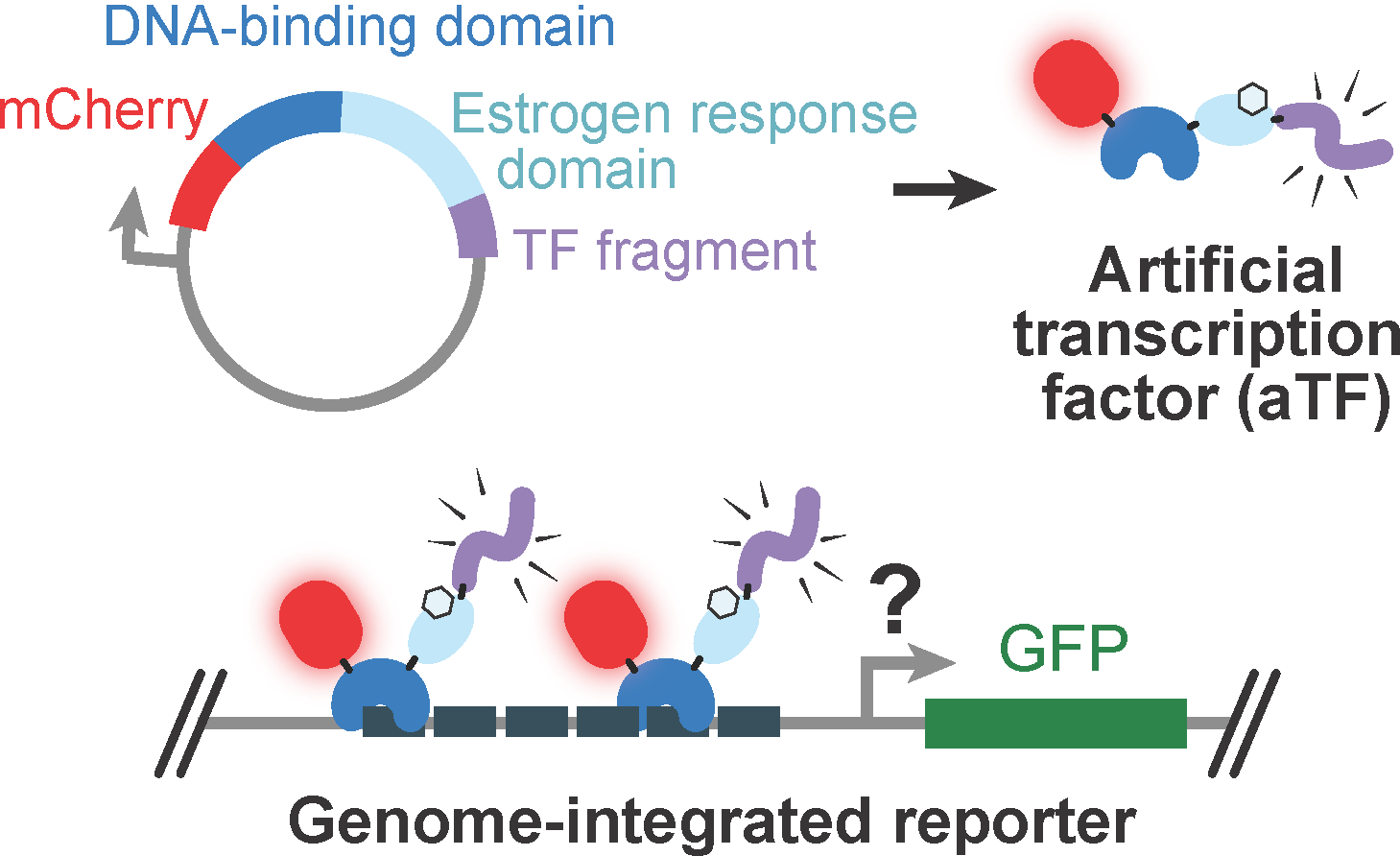
Modeling Transcription Factor-Coactivator Interactions
Stanford University School of Medicine, October 2018 - December 2020
Mentor(s): Dr. Roger Kornberg, Adrian Sanborn
The activation domains of transcription factors are poorly annotated, and the biochemical mechanisms by which they recruit co-activators such as Mediator remain poorly understood. We developed high-throughput assays to screen libraries of peptide fragments for transcriptional activation activity and co-activator binding. We identified all activation domains (ADs) in yeast transcription factors and trained an accurate deep learning model (PADDLE) to predict ADs in human and other yeast proteins. Our results reveal new insights into the fundamental mechanisms of eukaryotic transcription.
Paper (eLife, 2021) Preprint Code PADDLE
Experience
-
Gilead Research Bioinformatics Intern
Gilead Sciences, Foster City, Summer 2018
Mentor(s): Dr. David Lopez
As an intern on the Research Bioinformatics team, I used gene coexpression networks and transcription factor motif enrichment to study biological mechanisms of non-alcoholic steatohepatitis (NASH) disease progression.
Projects
-
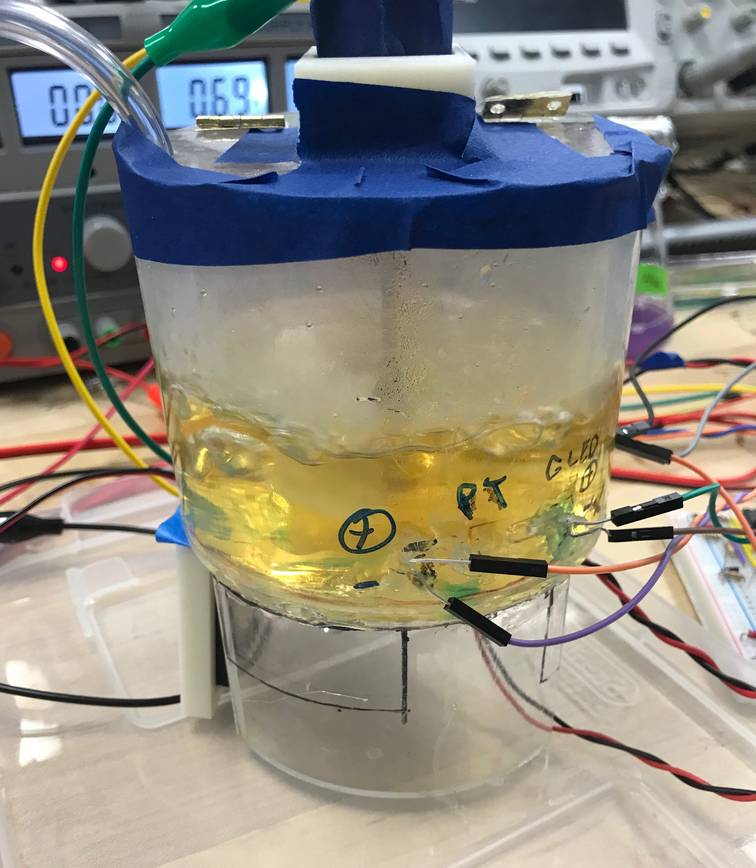
Web-controlled E. coli Fermenter
Stanford BIOE 123, Winter 2018
Team: Michael Becich, Augustine Chemparathy
Mentor(s): Dr. Ross Venook, Dr. Kwabena Boahen
We designed and built a fermenter that could be remotely controlled and monitored. Adjustable settings included heat, stirring velocity, and air flow rate; data measurements included optical density and temperature. The user could manually control the system or provide set points for automatic operation.
Code Video
The physical fermenter was built out of machined plastic and common electrical components (a detailed list is provided on the project repository). We used an Arduino Micro microcontroller to control the fermenter. A Wemos D1 mini WiFi microcontroller connected to the Arduino Micro was linked to Stanford’s network to locally host a webservice. -

Modeling and Prediction of Drug Toxicity from Chemical Structure
Stanford CS 221, Fall 2017
Team: Joyce Kang, Rifath Rashid
Mentor(s): Anna Wang
Humans are exposed to many different chemical compounds throughout the life course from sources including food, cleaning products, and drugs. In 2014, the Tox21 data challenge was launched by the National Center for Advancing Translational Sciences (NCATS) at the U.S. National Institutes of Health (NIH) to better understand the potential of compounds to disrupt biological pathways in possibly toxic ways. The Tox21 library comprises over 10,000 chemical compounds, and data was generated from biological assays measuring each compound’s effect on nuclear receptor signaling and cellular stress pathways.
We trained a 5-layer neural network, achieving a relatively high accuracy of 0.79 and AUC of 0.78 on the validation set. Additionally, by computing the gradient of prediction probabilities with respect to out input features, we were able to recover insights about structure-activity relationships. Carbonyl groups (e.g, ketones, esters, and carboxylic acids) were most predictive of toxicity, while the presence of many aromatic rings tended to be a negative predictor of toxicity.
Code Poster
